Quick start guide for vacamole
quick_start_guide.RmdOverview
vacamole is an R package that features a deterministic age-structured compartmental susceptible-exposed-infectious-recovered (SEIR) model and supporting functions. The model was developed to determine the impacts of different COVID-19 vaccination strategies on disease outcomes; therefore the model is an extended SEIR model to include states for severe disease outcomes and vaccination status.
Note: this package and vignette are still under development.
Model description
The model is a deterministic compartmental SEIR model. The population is partitioned into nine 10-year age groups (0-9, 10-19, …, 70-79, 80+). Within each age group we further partition the population into those who are unvaccinated, vaccinated with 1 dose, or vaccinated with 2 doses and then finally into disease states: susceptible (S), infected but not yet infectious (E), infectious (I), hospitalized (H), in intensive care (IC), return to the hospital ward after intensive care (HIC), recovered (R), and dead (D) (Figure 1).
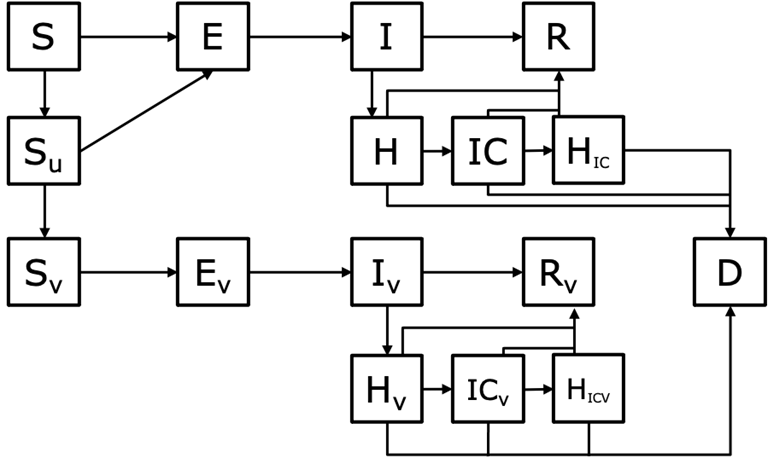
Figure 1. Basic conceptual model diagram. This diagram does not include the additional states after the second dose of vaccination or the age structure in the model. S = susceptible, E = exposed, I = Infectious, R = Recovered, H = hospitalized, IC = In intensive care, HIC = return to the hospital ward following treatment in IC, Su = vaccinated, but not yet protected, D = dead. States with subscript V indicate individuals who are vaccinated and protected by vaccination. This model assumes the “leaky” vaccine protection, so vaccinated and protected individuals can still be infected, hospitalized, etc. but at a reduced rate.
We assume that individuals who recover from infection cannot be re-infected. When a person is vaccinated, they first enter a hold state where they are vaccinated, but not yet (fully) protected (Shold1d or Shold2d). After a delay period, they enter the vaccinated and protected state for the dose they have received (Sv1d or Sv2d). We assume that vaccination only affects susceptible individuals.
The model is designed to incorporate a single vaccine product with a 2-dose regimen that 1) reduces susceptibility to infection, 2) reduces risk of hospitalization if a vaccinated individual is infected, and 3) reduces risk of infecting others (transmission) if a vaccinated person is infected. The vaccine is assumed to provide “leaky” protection, which means that the vaccine reduces the probability of transitioning between states for each vaccinated person. For example, a vaccine with 70% effectiveness against infection after the first dose would reduce the probability of transitioning from Sv1d to Ev1d by 70% compared to someone who is unvaccinated (moving from the S compartment to the E compartment). However, since there is more than one vaccine product currently licensed for use against COVID-19, we incorporate different vaccine products by taking the daily weighted average of the number of people with each vaccine product (and dose), the corresponding delay to protection of each vaccine product, and the vaccine effectiveness against each outcome. Rate of vaccination for each day, vaccine product, and dose for each age group are model inputs.
Vignette Outline
This quick start guide is designed to provide the basic instructions to use vacamole. The following steps are covered in this guide:
- Installing vacamole
- Data sources
- Specifying inputs
- Running the model
1. Install vacamole
The easiest way to install the development version of vacamole is to use the devtools package:
devtools::install_github("kylieainslie/vacamole")
#> rlang (0.4.11 -> 0.4.12) [CRAN]
#> cli (3.0.1 -> 3.1.0 ) [CRAN]
#>
#> There are binary versions available but the source versions are later:
#> binary source needs_compilation
#> rlang 0.4.11 0.4.12 TRUE
#> cli 3.0.1 3.1.0 TRUE
#>
#>
checking for file 'C:\Users\ainsliek\AppData\Local\Temp\RtmpgxUMB7\remotes59705a31534e\kylieainslie-vacamole-89e0bdc/DESCRIPTION' ...
v checking for file 'C:\Users\ainsliek\AppData\Local\Temp\RtmpgxUMB7\remotes59705a31534e\kylieainslie-vacamole-89e0bdc/DESCRIPTION'
#>
- preparing 'vacamole': (693ms)
#> checking DESCRIPTION meta-information ...
checking DESCRIPTION meta-information ...
v checking DESCRIPTION meta-information
#>
- checking for LF line-endings in source and make files and shell scripts
#>
- checking for empty or unneeded directories
#>
Omitted 'LazyData' from DESCRIPTION
#>
- building 'vacamole_1.0.tar.gz'
#>
#>
library(vacamole)
# Load other packages
library(dplyr)
library(tidyr)
library(readxl)
library(kableExtra)2. Data sources
vacamole contains several example data sets in the data/and inputs/ directories that can be used to run through the package functions. The example data sets include 1. Daily confirmed cases of COVID-19 from the Dutch national notification database Osiris from 1 January 2020 to 22 June 2021 RIVM Open Data. 2. Synthetic vaccination distribution schedule. 3. Contact matrices from the Pienter Corona study prior to the COVID-19 pandemic and in April 2020, June 2020, September 2020, February 2021, and June 2021.
3. Specifying inputs
vacamole expects several inputs namely, a named list of parameters for the initial number of individuals in each compartment, the rate of transitioning between states, contact matrices, and vaccination schedule. The following section will walk the users through creating each of these inputs. Some inputs values are stored in the inputs directory. A table of the required inputs is shown in Table 1. ## Vaccination schedule A synthetic vaccination schedule is provided in the inputs directory. The vaccine schedule should have columns for each vaccine type and dose by age group. The rows should be the proportion of each age group vaccinated with the vaccine dose and type per time period (usually day). The model expects four different vaccine types. For single dose vaccine regimens, the columns for the second dose are set to zero.
| date | pf_d1_1 | pf_d1_2 | pf_d1_3 | pf_d1_4 | pf_d1_5 | pf_d1_6 | pf_d1_7 | pf_d1_8 | pf_d1_9 |
|---|---|---|---|---|---|---|---|---|---|
| 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2020-01-07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 |
| 2020-01-08 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 |
| 2020-01-09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 |
| 2020-01-10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.4 |
| 2020-01-11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5 |
| 2020-01-12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.6 |
| 2020-01-13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.7 |
| 2020-01-14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0.75 |
The vaccination schedule needs to be converted before input into the model. The vaccination schedule data frame is converted into a list with weighted averages of the following quantities: vaccination rate, vaccine effectiveness, and delay to protection based on the number of people in the population who receive each dose and type of vaccine at each time point.
# read in vaccination schedule
vac_schedule <- read_xlsx("../inst/extdata/inputs/vac_schedule_18plus_synth.xlsx", col_types = c("date", rep("numeric", 80))) %>%
select(-starts_with("X"))
# read in vaccine effectiveness parameters
# these will be used when converting the vaccine schedule into weighted vaccine effectiveness, delay to protection, and vaccination rate
ve_info <- readRDS("../inst/extdata/inputs/ve_params.rds")
# convert vaccination schedule
# the vaccination schedule is assumed to be cumulative over time
vac_schedule1 <- convert_vac_schedule(
vac_schedule = vac_schedule,
ve = ve_info[[1]],
delay = ve_info[[2]],
hosp_multiplier = ve_info[[3]],
ve_trans = ve_info[[4]]
)Parameter values
# read in transition rates
transition_rates <- readRDS("../inst/extdata/inputs/transition_rates.rds")
# read in transmission matrix
april_2017 <- readRDS("../inst/extdata/inputs/contact_matrices/converted/transmission_matrix_april_2017.rds")
# define population size (by age group)
age_dist <- c(0.10319920, 0.11620856, 0.12740219, 0.12198707, 0.13083463,0.14514332, 0.12092904, 0.08807406, 0.04622194)
n <- 17407585 # Dutch population size
n_vec <- n * age_dist
# create named list of parameter
params <- list(beta = 0.0004,
beta1 = 0.14,
gamma = 0.5,
sigma = 0.5,
epsilon = 0.01,
N = n_vec,
h = transition_rates$h,
i1 = transition_rates$i1,
i2 = transition_rates$i2,
d = transition_rates$d,
d_ic = transition_rates$d_ic,
d_hic = transition_rates$d_hic,
r = transition_rates$r,
r_ic = transition_rates$r_ic,
p_report = 1/3,
c_start = april_2017,
keep_cm_fixed = TRUE,
vac_inputs = vac_schedule1,
use_cases = TRUE,
no_vac = FALSE,
t_calendar_start = yday(as.Date("2020-01-01")),
beta_change = NULL
)Initial conditions
The model is partitioned into nine age groups, so users need to supply the initial number of individuals in each age group for each compartment. The initial conditions should be a named list. Below we start with a fully susceptible population and one infectious person.
# Specify initial conditions ---------------------------------
empty_state <- c(rep(0, 9)) # vector of zeros
init <- c(
t = 0,
S = c(n_vec[1:4], n_vec[5]-1, n_vec[6:9]),
Shold_1d = empty_state,
Sv_1d = empty_state,
Shold_2d = empty_state,
Sv_2d = empty_state,
E = empty_state,
Ev_1d = empty_state,
Ev_2d = empty_state,
I = c(rep(0,4),1,rep(0,4)),
Iv_1d = empty_state,
Iv_2d = empty_state,
H = empty_state,
Hv_1d = empty_state,
Hv_2d = empty_state,
H_IC = empty_state,
H_ICv_1d = empty_state,
H_ICv_2d = empty_state,
IC = empty_state,
ICv_1d = empty_state,
ICv_2d = empty_state,
D = empty_state,
R = empty_state,
Rv_1d = empty_state,
Rv_2d = empty_state
)
# create vector of time points
times <- seq(0,nrow(vac_schedule)-1, by = 1)3. Run model
The model is coded as a system of ordinary differential equations that are numerically solved using the lsoda function in the deSolve package.
# Solve model ------------------------------------------------------
seir_out <- lsoda(init, # initial conditions
times, # time vector
age_struct_seir_ode, # model function
params) # parameters
# post-process the model output ------------------------------------
seir_out <- as.data.frame(seir_out)
out <- postprocess_age_struct_model_output(seir_out)